DM-CANCER
Data Mining for Carcinogen Patterns
From the Data-Centric Computing group and Computational Genomics group @ BSC
Introduction of the project and main goals
Data Mining on Carcinogen Rearrangements (CR) attempts to address the problem of finding patterns and relevant/useful information and knowledge from CR graph representations, involving pattern mining and sub-graph isomorphism and coloring problems, known to be NP-hard. The problem consists in identifying CR patterns across samples, with statistical relevance as tumor root cause or consequence in front of random noise, which would allow identifying potential markers with clinical value. DM-CANCER addresses the mining problem by representing data as graphs to apply heuristic methods form mining data, enhanced by Machine Learning methods like Neural Networks, used as generative models for prediction and forecasting of pattern components, also for learning multi-dimensional structured data. In addition, DM-CANCER proposes the use of computational distributed solutions for the mining process leveraging the most advanced supercomputing facilities.
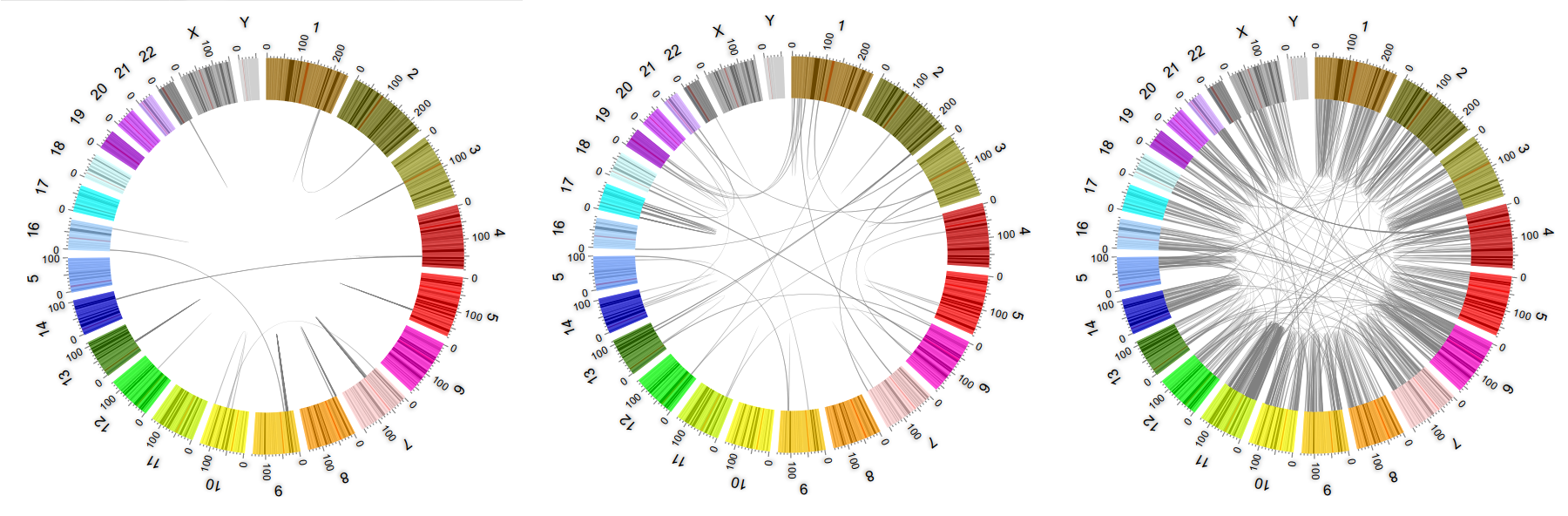
DM-CANCER is composed by BSC researchers from the Computer Sciences department and the Life Sciences department
The focus of the group is to accelerate the processing of data-driven workloads, including large analytics as well as stream processing, in heterogeneous execution frameworks. The goal of the group is to advance in the field of methods, mechanisms and algorithms for the management of heterogeneous data-centre workloads.
The focus of the group is to decipher and understand the biology of genomes. Genomes, as the molecules of life, contain the necessary information for the development and evolution of living organisms. Despite this information is encoded in a four-letter code; it presents different levels of coding possibilities and complexity
Copyright © Barcelona Supercomputing Center, 2022 - All Rights Reserved - DM-CANCER